BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material
BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material: BSc is a three-year program in most universities. Some of the universities also offer BSc Honours. After getting enrolled for BSc, there are certain things you require the most to get better grades/marks in BSc. Out of those, there are BSc Study Material, BSc Sample Model Practice Mock Question Answer Papers along with BSc Previous Year Papers. At gurujistudy.com you can easily get all these study materials and notes for free. Here in this post, we are happy to provide you with BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material.

BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material
The unit of measurement most often used for microorganisms is the micrometer-a unit of length equal to one millionth (micro-) of a meter i.e. 10-6 m. The word is symbolized by the um-Greek letter u together with the abbreviation for meter, m. It is also equal to a thousandth of a millimeter i.e. 10-3 mm. The figure shows some common metric equivalents used in microbiology. It may be seen that the cells of the microbes listed here, range in size from the large, almost-visible protozoa, measuring 100 um to the incredibly small viruses at about 0.01 um, ten thousand times smaller. Mold filaments are fairly long and visible to the naked eye.
However, their individual cells measure approximately 40 um by 10 um. Unicellular fungi, the yeasts, commonly have cells of 25 um in diameter. The largest bacteria are about 20 um in length while the smaller members of this group, the rickettsiae may be about 0.5 um long, and the chlamydiae, about 0.20 um.

The viruses are too small to be seen by conventional light microscope techniques. Their measurements vary from the large pox viruses at 0.25 um to the polio viruses at 0.02 um. Virologists use the term nanometer, a unit of length equal to one billionth (nano-) of a meter. A nanometer (nm) is a thousandth of a micrometer; therefore 0.02 um is equivalent to 20 nm. A nanometer is also the unit used in measuring the wavelength of radiant energy such as visible or ultraviolet light.
The introduction of a light microscope could make it possible to determine the overall shape of cells and their arrangement. Moreover, some internal structures could be seen by using phase-contrast or interference microscopy or by staining methods with bright-field or fluorescence microscopy. Most of the stains in current use, such as basic dyes are relatively non-specific and thus gave little idea of the chemical nature or function of a cellular component. Some are more specific: fat-soluble dyes such as Sudan Black stains lipids, iodine colours staro granules, and acridine orange fluoresces with nucleic acids.
However, in small microbes like bacteria, the cell size (about 0.5 um diam.) is only slightly greater than the theoretical limit of resolution of the light microscope (about 0.2 um). The study of the ultra-structure of cells, therefore, depended upon the transmission and stereo scan electron microscopy. Unfortunately, one of the main problems in electron microscopy is in the interpretation of the chemical nature and function of the structures seen. There are only a few specific stains available.
Of course, there may be potential in the current use of specific antibodies labeled with electron-dense components like ferritin or colloidal gold, but there are technical difficulties in their use.
Consequently, the primary method used for relating a given structure to function is to break open the cell and fractionate the components which are then subjected to biochemical analysis. There may be some problems in rupturing a cell without denaturing the more sensitive components, particularly in bacteria with their tough walls and small size. However, in recent years methods of breakdown could be developed and most of the major cell structures have been isolated in a reasonably pure state in which they perform their functions normally. In bacteria, for instance, there are following two methods of cell breakdown.

Mechanical disruption. Violent shaking of a bacterial suspension with glass beads causes rupture of the cell walls and liberation of the cytoplasmic contents. The walls can be concentrated by centrifugation and treated with appropriate solvents and enzymes to remove contaminating components. The wall isolated in this way retains the shape of the original cell and is also termed a wall ghost. In some bacteria, the wall can also be solubilized completely by the enzyme, Lysozyme, a component of many animal secretions and fluids.
Lysozyme treatment. Exposure of a sensitive bacterial species to lysozyme normally causes complete cell lysis. However, if the treatment is given in the presence of an osmotic stabilizer such as an isotonic sucrose solution, the cell assumes a spherical shape as the wall is solubilized.
The product after the removal of the wall is called a protoplast that is bounded by a cytoplasmic membrane. The following points could emerge from such experiments:
(1) The wall determines cell shape since the protoplast is spherical irrespective of the original shape.
(2) The wall is responsible for the mechanical strength of the bacterial cell since a protoplast is very prone to mechanical or osmotic lysis while the cell is not.
(3) The cytoplasmic membrane and not the cell wall is responsible for the selective permeability of the cell surface; this remains unchanged in the protoplast. Further studies have shown that the protoplast retains most of the enzyme systems of the cell and is capable of regeneration and growth whereas the wall is not.
If a protoplast is subjected to controlled osmotic lysis by a gradual reduction in the external osmotic pressure, the cytoplasmic membrane ruptures liberating the cytoplasm and its components. The membranes and the components of the cytoplasm can then be isolated by differential centrifugation. These remain unaffected by mild lytic treatments.
BSc Structure of Microorganisms in Microbiology Notes Study Material
With the help of such methods, the structure and function of the components of apparently typical cells have been studied. It has become clear that there are two basic types of cells – prokaryotic and eukaryotic. The former is restricted to microorganisms (bacteria, including blue-green or cyanobacteria), whereas the latter occurs in microorganisms (fungi, protozoa, and algae) and in animals and plants. There are also viruses that have a simple acellular structure. The prokaryotes and eukaryotes share the common basic unit of life, the cell. All cells have the following essential components:
(1) DNA, as genetic information for replication.
(2) RNA, for protein synthesis.
(3) Enzymes, for catalysis.
(4) A membrane, to maintain the internal environment.
(5) Cytoplasm, as a solvent for the utilization of food.
Thus, all cells share a common chemical composition, the common chemical activities of metabolism, and a common physical structure of organization, and together these give the potential for growth and self-replication.
Cells: The Basic Organisational Units of Living Systems
We consider briefly the properties of the structural and functional subunit of all living organisms – the cell. A cell is a self-contained unit separated from its surrounding by a membrane that serves as a limiting boundary. The membrane regulates the flow of materials into and out of the cell allowing the critical maintenance of the cell’s internal contents in a more highly organized state than the cell’s external environment. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
Microorganisms can be classified on the basis of their cellular organization. It may be seen that some microbes as viruses, viroids, and prions lack cell walls (acellular). They will be discussed in detail with a separate topic. Within their host cells, they are capable of acting as live systems because the host cell provides the essential boundary membrane. In contrast to these, cellular microbes, all other living organisms are composed of either prokaryotic or eukaryotic cells.

We would describe below in somewhat detail the various structures associated with typical prokaryotic and eukaryotic cells. These include the following:
(1) Structures involved with the movement of cells – Flagella, Cilia.
(2) Structures involved in the attachment of cells – Glycocalyx, Pili, Fimbria, etc.
(3) External structures that protect the cells – Cell wall, Capsule, Slime layers.
(4) Structure involved in the regulatory movement of materials into and out of cells – Cytoplasmic (cell) membrane.
(5) Sites of energy transformation where ATP is generated – Cytoplasmic membrane, Internal photosynthetic membranes, Chloroplast, Chromatophores, Mitochondria.
(6) Structures involved in information flow in cells – Ribosomes.
(7) Cellular storage of genetic information – Bacterial chromosome, Plasmids, Nucleus, and Chromosomes.
(8) Structures involved in coordinated material movement and storage – Exoenzymes, Endoplasmic reticulum, Golgi apparatus, Lysosomes, Microbodies, Vacuoles, Cytoskeleton, Inclusion bodies.
(9) Structures involved in survival during adverse environmental conditions – Endospores in bacteria, Various types of spores in eukaryotes.
Prokaryotic Microorganisms
We would prefer to refer to all these organisms as bacteria. Although prokaryotes are often subdivided, such a division appears to be artificial in the light of the present state of knowledge about these microorganisms. Let us consider the range of prokaryotic microbes in terms of their structure.
Cell shapes. Various cell shapes occur in prokaryotes: spheres or coccoid bacteria, rods, helically grown cells (spirillum-type organisms), comma-shaped (vibrios), box-and plate-shaped, filaments, bacteria with stalks and prosthesis, and branched systems. There are three basic cell shapes: the sphere (coccus); the rod (bacillus); a cylinder with rounded ends; and the curved rod, either as one slight curve (vibrio) or as a spiral or helix. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
Multicellular structures. Bacteria basically are unicellular. However, sometimes cell division and cross-wall formation are not followed by the separation of the daughter cells. In this way, an undifferentiated multicellular structure is produced. The shape of this multicelled structure depends upon the planes of cell division.
The rods always divide in one plane, and if the cells remain attached a multicellular filament is formed.

In spherical cells, however, a variety of shapes can occur depending on the plane of cell division. These are as follows:
(i) cells divide in one plane and remain predominantly attached in pairs, e.g. Diplococcus
(ii) cells divide in one plane and remain attached to form chains, e.g. Streptococcus.
(iii) cells divide into two planes to give plates, e.g. Pediococcus.
(iv) cells divide into three planes regularly to produce a cube, e.g. Sarcina
(v) cells divide into three planes irregularly producing bunches of cocci, e.g. Staphylococcus.
Spiral bacteria are predominantly unattached but the individual cells of different species show striking differences in length and in tightness of spiral.
In all these instances, the multicellular form is made up of separate individual cells. However, in actinomycetes, multi-nucleate cells without cross walls produce branched mycelia of indefinite length which appear superficially similar to those of filamentous fungi. Various types of multicellular forms are-
Stalk. A few bacteria have a stalk by which they attach to a solid substratum using a holdfast at the tip. Upon cell division a flagellate cell is produced which swims around, eventually settling on a new surface where it forms a stalk in place of the flagellum. This life cycle is very simple having an attached and a swarming phase. For example, Hyphomicrobium.
Buds. Most bacteria divide by symmetric division into two equal halves. However, a few as Rhodomicrobium divide by budding as in yeasts. The growth is polarised and cell division is asymmetric.
Motility structures. Motility may be by flagella, by gliding, or by axial filaments. We shall refer to these structures later.
Spores and cysts. We shall describe them in the following pages.
Fine Structure of Prokaryotic Cell
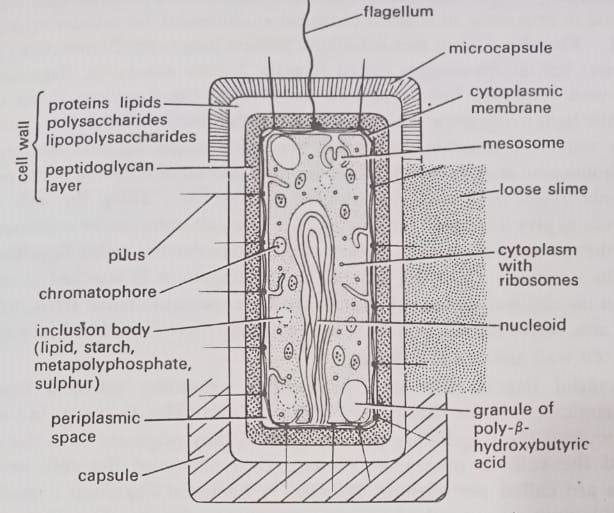
The prokaryotic cells vary in size from a Mycoplasma (a sphere of about 0.12 um diameter) to a blue-green bacterium like Oscillatoria (a rod of dimensions as much as 40×5 um), but the majority have a diameter in the region of 1.0 um. The components of a typical prokaryotic cell are shown in Figure. These are as follows:
Cytoplasm or cytosol. The cytoplasm is sequestered within the cell membrane. Does cytosol has a structure or is it just like a solution of enzymes and metabolites in a test tube? Whether cytosol in vivo is just a disordered solution (a “bag of enzymes”) or indeed highly organized (i.e. structure) is a matter of some debate. Though there is evidence that it has structure but the concept is still poorly understood.
In an electron microscope, it is not organized. It is concentrated and contains a variety of enzymes, coenzyme, and metabolites, perhaps in the form of aqueous fluid or semifluid ground substance or matrix. The matrix is a complex mixture containing in solution a variety of inorganic ions, amino acids, some proteins, lipocomplexes, peptides, nitrogenous bases, sugars, vitamins, enzymes, coenzyme, etc. Its main function is in intermediary metabolism and in providing an equable chemical environment for cellular activities.
Flagella. Most motile bacteria possess long (up to 20um), thin (20 nm diameter) helical appendages called flagella. Unlike the eukaryotic flagellum, the prokaryotic flagellum has no definite membrane. The flagellum is not visible using the light microscope without increasing its effective diameter by coating it with a suitable precipitate. In the electron microscope, negative-staining with phosphotungstic acid shows that the flagellum is made up of identical (three or more in number) sub-units arranged (intertwined) helically along the axis of the flagellum to give a hollow tube. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
These sub-units can be separated from each other by acidification and consist of protein molecules called flagellin. Each flagellin subunit is about 4.5 nm thick. The flagellum is attached at one end through the cell wall to the cell membrane by a special terminal hook, which in turn is attached to the basal body. The rings of the basal body are attached to the cell wall and cell membrane.

Bacterial flagella are long projections extending outward from the cytoplasmic membrane that propel bacteria from place to place. In some, such as Pseudomonas, the flagella are polar flagella as they originate from the end or pole of the cell. In others, such as Proteus, they surround the cell, and such flagella are called peritrichous flagella. A bacterial flagellum consists of a single filament composed of protein (flagellin) subunits.
The characteristic hook structure of the flagellum allows it to spin like a propeller and thereby propel the bacterial cell. Effectively, the structure allows the flagellum to spin like the shaft of an electric motor. Rotation of the flagellum requires energy which is supplied by a hydrogen ion gradient across the cytoplasmic membrane.
BSc Structure of Microorganisms in Microbiology Notes Study Material
The arrangement and number of flagella on a cell can be useful criteria for identification and classification. The following four types of flagellation patterns are common in bacterial cells, as shown in Figure.

(a) Monotrichous. There is a single flagellum at one pole of the cell.
(b) Lophotrichous. There are several or numerous flagella at one pole.
(c) Amphitrichous. The cell bears at least one flagellum at each pole.
(d) Peritrichous. There are flagella all over the surface of the cell.
The flagellum rotates, being driven by a rotary motor in its basal body. The hook acts as a universal joint, and the flagellum acts similarly to a ship’s propeller, projecting backward and driving the cell forward by exerting a viscous force against the aqueous medium.

The function of flagella is in locomotion and all naturally occurring flagellate bacteria are motile. However, some other less common types of motility are also shown by some prokaryotes. They have helical cells with axial filaments like flagella.
Specialized movement in bacteria
Chemotaxis
This is the mechanism for swimming toward or away from chemical stimuli, a behavior known as chemotaxis. The chemosensors in the cell envelope, called binding proteins detect certain chemicals and signal the flagella to respond. Bacteria have a memory system that allows them to compare the concentrations of chemicals as they swim along so that they effectively detect chemical concentrations over distances many times the length of a cell.

BSc Structure of Microorganisms in Microbiology Notes Study Material
To understand how chemotaxis works, we need to recognize that when bacteria move, they periodically change direction rather than reaching their destination by swimming in a single straight line. The straight-line movements are known as runs, and the turns – which occur when the bacteria stop – are called tumbles or twiddles. At least in those with peritrichous flagella, the counterclockwise rotation of the flagellum results in a run and the clockwise rotation in a twiddle.
The direction of flagella rotation, and hence the length of a run – that is, the amount of time before the organism stops and tumbles – is determined by the interactions of chemosensors in cytoplasmic membrane with attractants or repellents (i.e. the chemicals). An increasing concentration of an attractant, for example, interacts with the chemosensors to decrease the frequency of tumbling, whereas a decreasing concentration of attractant causes increasing tumbling and hence shorter runs. The same is true for the interactions with repellents.
The net effect of this process, called a random walk, is a biased movement toward an attractant or away from a repellent based on the relative proportion of running and tumbling. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
Magnetotaxis
Some bacterial cells contain the inclusion of iron granules known as magnetosomes, which permit them to orient their movement in response to magnetic fields, a phenomenon known as magnetotaxis. Bacteria can use these granules to navigate along the earth’s magnetic field. Some bacteria move predominantly north and others the south. Magnetotaxis allows some anaerobic bacteria to orient themselves so that they point downward into the sediment. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
Phototaxis
Some bacteria are able to detect and respond to differences in light intensity, a phenomenon called phototaxis. In some bacteria, this is similar to chemotaxis. Other bacteria form membrane-bound gas vacuoles that enable them to respond to light. Boundary layers of these vacuoles are not true membranes but are composed exclusively of proteins, which are hydrophilic as well as hydrophobic.
In aquatic bacteria, gas vacuoles provide a mechanism for adjusting the buoyancy of the cell and thus the height of the bacterium in the water column. Many aquatic cyanobacteria use their gas vacuoles to move up and down in the water column, depending upon light intensity levels, to achieve optimal conditions for photosynthesis.
Pili, fimbriae and spinae. The term pili were introduced by Brinton (1959) and fimbriae by Duguid et al (1955). Both are non-flagellar appendages of the cell. They are extremely fine, non-flagellar appendages which look superficially similar to flagella. Pilis has been observed mostly in Gram-negative rods. They measure 3-25 nm in diam, and 0.5-20 um in length. Pilis is also made up of individual protein sub-units of pilin, arranged helically to form filaments. However, they differ from flagella in the following respects:
(i) the filament is usually straight and is shorter than a flagellum.
(ii) the diameter is smaller (about 10 nm)
(iii) the function is not in motility.
In some bacteria, there are longer, the f-pili on male cells that act as a bridge between conjugating cells. The smaller pili perhaps serve for the attachment of cells to their substrates, e.g. pathogens to their hosts, for example, Neisseria gonorrhea to cells of the human urinary tract. Both flagella and pili are antigenic and can be specific sites of attachment for bacteriophages. Pili are plasmid-coded and the plasmid is said to code for conjugation and resistance to antibiotics, colicin, or hemolysin production. Fimbriae are involved in adhesion, between themselves as well as between them and the eukaryotes.
In some Gram-positive bacteria spine have been reported. These are tubular, pericellular non-prosthecate rigid appendages made up of a single protein moiety – spin-in. They are said to help adjust cells to some environmental conditions such as salinity, pH, temperature, etc.
Many bacterial cells are surrounded by a specialized structure called glycocalyx that plays a role in the attachment process. This is a mass of tangled fibers of polysaccharides or branching sugar molecules surrounding an individual cell or colony of cells. It may act to bind cells together, forming multicellular aggregates. Additionally, in some bacteria glycocalyx is also involved in attachment to solid surfaces.
Some pathogenic bacteria adhere to animal tissues they invade via a glycocalyx. In aquatic habitats, bacteria seem to be held to rocks through the slime layers they secrete. An extensive polysaccharide slime, dental plaque enables bacteria to adhere to the tooth.
Capsule. Some prokaryotes have a gel layer called the capsule surrounding the cell wall. It can be seen in the light microscope by negative staining with a dye like Indian ink. In an electron microscope, it normally looks like an amorphous shrunken layer. They are synthesized by the cell membranes. The gel is usually formed of a polysaccharide (1-2% dry wt.) in water and there is a wide variety of different monosaccharide components joined in many different ways. Occasionally, there may be polypeptides also, as the peculiar polymer of the unusual D-glutamic acid found in anthrax-causing bacteria.
The capsule serves mainly as a protective layer against attack by phagocytes and by viruses; it may also help to prevent too rapid and lethal a loss or gain of water in the recurrent dehydration and hydration that occurs in many habitats. Finally, the capsule usually has an ion-exchange capacity which may aid in the concentration and uptake of essential cations.
Cell wall. The cell wall is the dense layer surrounding the cell membrane. The main function is to provide a mechanically strong bonding layer. Some, such as Mycoplasma, do not have a wall and thus exist only in restricted habitats.

The cell wall is not semi-permeable membrane but it can act as a molecular sieve preventing large molecules from passing through. In Gram-negative cells, some enzymes and metabolites are trapped between the cell membrane and the outer membrane of the cell wall to form the periplasm. The wall components of the cell can be strongly antigenic.
BSc Structure of Microorganisms in Microbiology Notes Study Material
All peripheral layers including the cytoplasmic membrane, which enclose the cytosol (cytoplasm) are designated as cell envelopes. The cell wall is structurally and chemically a distinct part of the cell. The rigidity and strength of cell walls are mainly due to strong fibers composed of heteropolymers generally called peptidoglycans or mucopeptides but also referred to as glycopeptide, muropeptide, glucosamine-peptide, much complex, murein, etc.
These fibers form a relatively coarse (at the molecular level), three-dimensional, tough meshwork rather than a solid structure. These fibres offer no resistance to inward movement of water, food as minerals, glucose, amino acids. All wastes of cells can pass outward.
The peptidoglycans consist of alternating units of N-acetyl-glucosamine and N-acetyl muramic acid with B-1, 4-linkages. This backbone structure appears to be the same in all prokaryotes studied. Linked to the muramic acid is a short peptide that varies in composition but always contains a minimum of three amino acids, viz. alanine, glutamic acid, and either diaminopimelic acid (DAP) or lysine.
Glutamic acid and one of the alanines of peptidoglycans are in D-isomeric form rather than L-configuration which is characteristic of amino acids found in proteins. Muramic acid, DAP, and several D-aminoacids are found only in association with prokaryotes. In this way, one of the most important aspects of the chemical structure is the presence of unique monomers in both the polysaccharide component (N-acetyl-muramic acid) and the polypeptide component (D-aminoacids and sometimes diaminopimelic acid).

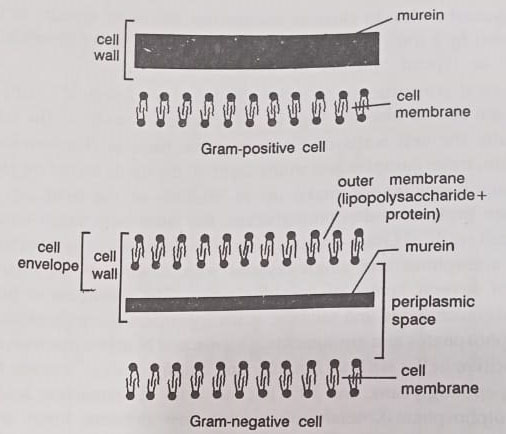
The structural rigidity of the peptidoglycans is achieved by cross-linking these polymers. The type and extent of cross-linkage vary among different species. In some bacteria, a short peptide (e.g. a pentaglycine) is used to link the tetrapeptide side chains extending from the muramic acid units, while in others, the terminal D-alanine of one tetrapeptide may be covalently linked to the DAP of an adjacent tetrapeptide. The peptidoglycans of Gram-positive bacteria are more extensively cross-linked than those of Gram-negative.
The peptidoglycans alone do not make the cell wall itself. In fact, cell walls are structurally and chemically very complex. Electron micrographs of thin sections of typical Gram-negative bacteria often show a multilayered (as many as five) cell wall. The different layers as seen under an electron microscope are the periplasmic space, the peptidoglycan layer, a more or less structureless zone (the “intermediate zone”), the outer membrane, and lipopolysaccharides (Acker, 1977).
The specific features of Gram-negative bacteria cell walls are the presence of (i) periplasmic space or also called periplasmic gel (Hobot et al, 1984), about 3-4 nm wide (ii) an intermediate layer, located outside the peptidoglycan and defined as a gap between peptidoglycan and outer membrane. According to Hobot et al (1984), the intermediate layer does not exist and perhaps periplasmic gel might also fill this space. (iii) fair amounts of lipoproteins and (iv) the outer membrane, whose three major components are proteins, phospholipids, and lipopolysaccharides lines. (BSc 2nd Year Structure of Microorganisms in Microbiology Notes Study Material)
In an electron microscope, this layer appears as two dense lines separated by a transparent zone i.e. exhibiting an aspect identical to a “unit membrane” or typical “bilayer”.

Biochemical constituents of the cell wall of E. coli, a typical Gram-negative bacterium are shown also in Figure, a schematic model of the cell wall.

Generally, the cell walls of Gram-negative bacteria (Escherichia, Salmonella) are thin, more complex, and multi-layered; the lipids including phospholipids and lipopolysaccharides make up to 80-85% of the total cell wall and remaining proteins and peptidoglycans, the latter only single-layered. The cell walls of Gram-positive (Bacillus, Staphylococcus) bacteria tend to be thicker, amorphous, and single-layered with 20-80% of the total cell wall composed of several layers of peptidoglycans. The remainder is made up of proteins, polysaccharides, and teichoic acids (polymers made up of either ribitol or glycerol phosphates and amino acids, glucose, and N-acetyl glucosamine).
Thus in Gram-positive cells, such as Bacillus and Staphylococcus species, the wall is about 50% peptidoglycans, and 50% teichoic acids or lipoteichoic acids (polymerized polyol phosphates), teichuronic acids, few proteins, lipids, and neutral polysaccharides. The Gram-positive cell walls appear for the most part to be more homogeneous because distinct layers are not readily apparent and the peptidoglycan is distributed throughout the cell wall more uniformly in a three-dimensional network.
The detailed cell wall structure of E. coli, a Gram-negative rod is shown in Figure. The structural and chemical characteristics of Gram-negative and Gram-positive bacteria are presented in the Table and Figure.
Cell walls of archaebacteria
Cell walls of archaebacteria do not contain peptidoglycan (murein); rather their wall structures show great biochemical diversity. Some archaebacterial cell walls have pseudomurein which resembles peptidoglycan of eubacteria but contains N-acetyl-talosaminuronic acid instead of N-acetyl-muramic acid and lack the D-amino acids found in eubacterial cell walls. Other archaebacteria have walls composed of protein subunits; still, others have walls with different biochemical compositions. Although, variable in chemical structure, the walls are able to protect the cell membrane even in hot, acidic, and saline environments where many archaebacteria live.

Regular surface layers (S-layers). These are present external to the cell walls in all Gram-positive and Gram-negative bacteria even in those without peptidoglycans in their cell wall. Three main kinds of S-layers have been demonstrated in different cell envelopes of bacteria. S-layers are mainly composed of single homogeneous polypeptides with carbohydrates occasionally as a minor component. These layers have a predominantly acidic amino acid composition. These layers may act as a barrier or molecular sieve controlling the movement of external and internal factors such as toxic macromolecules. These may also protect the peptidoglycan from the action of lytic enzymes as lysozyme.
Cell membrane. This is the boundary layer of the protoplast and is about 6-8 nm thick. In thin sections, it appears a triple-layered structure consisting of two electron-dense (each about 2.5 nm thick) layers surrounding an electron-transparent one, a type of structure typical of most selectively permeable membranes in living organisms and called a unit membrane. The main chemical components are lipid and protein in about equal amounts in a fluid mosaic structure. This is composed of a bilayer sheet of phospholipid molecules, their polar heads on the surfaces, and their fatty-acyl chains forming the interior.
The protein components are embedded within the phospholipids, some spanning the membrane, some on one side, and some on the other. The membrane is fluid in that components can diffuse laterally in its structure. It is also asymmetric, and by its activities, it creates and maintains gradients across itself so that the inside of the cell is very different from the outside environment.
The prokaryotic cell membrane is a principal structural component of the cell, which is obvious from its following possible functions.
(a) Selectively permeable layer. It allows the entry and exit of some molecules but not others. This is very important, because if this barrier is broken down, essential metabolites may pass out of the cell, resulting in its death. The transport of molecules across the membrane in either direction usually involves their specific combination with protein molecules called permeases which are built into the membrane structure. Due to this specificity of combinations, a large number of different permeases may occur in any single cell.
(b) Energy production. The membrane is the site of electron flow in respiration and photosynthesis leading to phosphorylation (i.e. conversion of ADP to ATP) and therefore is the site of the enzymes and carriers in these reactions.
(c) Extracellular polymer production. The final stages in the synthesis of some of the polymers in the cell wall, capsule, and extracellular fluids are catalyzed by membrane enzymes.
(d) Site of chromosome attachment. The single chromosome and the cell membrane are attached at a specific point at which replication starts. The first stage in nuclear division involves duplication of this attachment site, followed by a progressive bidirectional replication of the DNA by two replication forks.

The cell membrane of archaebacteria. Archaebacteria have a cytoplasmic membrane with structures very different from true bacteria as well as eukaryotes and this feature distinguishes them from all other organisms. In an extreme environment which they inhabit, unusual physiologically specialized membranes are needed for survival. In contrast to eubacterial and eukaryotic cell membranes that contain straight-chain fatty acids linked to glycerol by ester linkages, the cell membranes of archaebacteria contain branched lipids.
The lipids are diethers in which a glycerol unit is connected by an ether link to phenols, the branched chains in which carbon atoms at regular intervals carry a methyl group. Moreover, glycerol has two optical isomers distinguished by the configuration of the molecule around the central carbon atom; the optical isomers rotate polarised light in opposite directions. Sulpholobus, an archaebacterium of high temperatures in acidic environments – has a cell membrane that contains long chain branched hydrocarbons twice the length of the fatty acids found in the cell membranes of eubacteria.
The lipid chains are long enough to extend from one side of the membrane to the other giving it the appearance of a monolayer while concealing its true bilipid structure. Similar unusual membrane structures occur in other archaebacteria of other extreme environments, including Thermoplasma living in high temperatures, and Halobacterium in habitats of high salt concentrations. The structure of these membranes makes them very resistant to conditions that would disrupt them and interrupt the function of a normal bilipid layer, enabling them to remain as semipermeable barriers in extreme environments.
Intracytoplasmic membranes. The cytoplasmic membrane may not exist as a simple structure underlying the cell wall and following its contours. However, infoldings may occur which can produce complex internal structures and increase their surface area. Consequently, there occur a number of morphologically and physiologically differentiated intracytoplasmic membranous structures in different prokaryotes. All such structures may be grouped into two main categories.
(a) Chromatophores. These are pigment-bearing membranous structures of photosynthetic bacteria. They occur in all Rhodospirillaceae, Chromatiaceae, and Cyanophyceae and vary in form as vesicles, tubes, bundled tubes, stacks, membranes, or thylakoids (as in cyanobacteria).
(b) Mesosomes. Mesosomes were earlier called the peripheral body or chondroid and have been seen in ultrathin sections of all Gram-positive and occasionally in Gram-negative bacteria. They have been assigned a number of functions by different persons from time to time. They have been defined as “vesicular, lamellar or tubular packets of membrane-enclosed by the invaginations of the plasma membrane” and “as seen in thin sections of whole or plasmolyzed cells the mesosome or its derived vesicles have all of the features of unit membrane profiles.”
According to some others, mesosomes are simply preparation artifacts. These are seen in chemoautotrophic bacteria with high rates of aerobic respiration, such as Nitrosomonas, and in photosynthetic bacteria such as Rhodopseudomonas, where they are the site of photosynthetic pigments. However, in electron micrographs of some bacteria, such as Bacillus licheniformis, there are seen some localized infoldings of the membrane, which may perhaps be the effect of chemical fixation on a specialized region of the cell membrane. Mesosomes a involved in septum formation, rather than being true structures in vivo.